SUPERRESOLUTION
Single-molecule microscopy
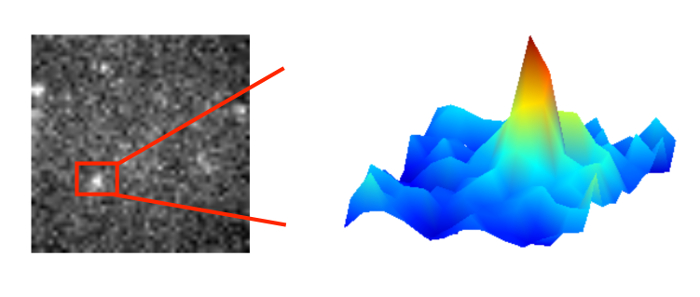
allows us to see dynamic proteins at work in living cells.
Super-resolved images
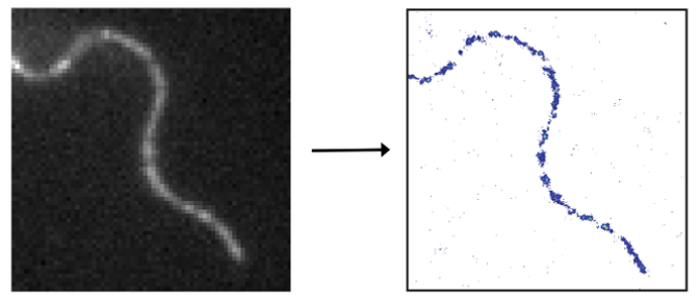
help to develop our understanding of the cellular nanostructure.
Highly motile cells
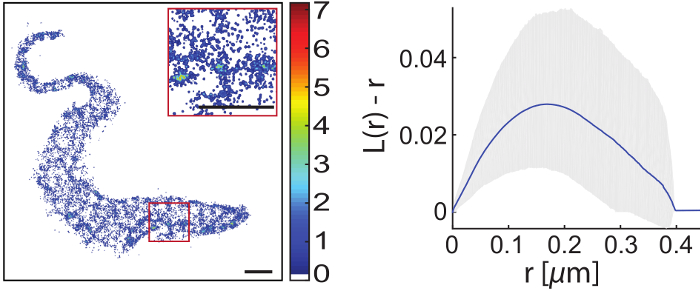
are accessible to super-resolution imaging after special treatment.
Resolution
In general, light microcopy is limited by diffraction. The light wave is diffracted by the lenses and mirrors of the microscope. As a result a fluorescent point object is imaged as an intensity distribution called Airy disk. The width of this disk is determined by the wavelength of light and the numerical aperture of the objective used. Usually, it is limited to 200-250 nm. If we aim to resolve two point objects close to each other, the width of the Airy disk defines the minimum separation required (Rayleigh criterion).
On the upside, the maximum of the Airy disk can be localized with considerable higher precision than the width of the disk. The localization precision of an isolated single molecule depends critically on the signal-to-noise ratio. Bright objects in a clean sample may be localized with a precision of down to a few nanometers.
Given the short de Broglie wavelength of high energy electrons, electron microscopy can achieve much better resolution. However, it is not suitable for live-cell imaging. In contrast, single-molecule fluorescence microscopy allows us to observe and track proteins at work in living cells. In the standard approach used in our lab, the temporal resolution is limited by the time to collect enough photons from a single fluorophore and the camera read-out. Frame rates up to 100 Hz are achieved.
In order to compute a super-resolved image of a densely-labeled structure, the fluorescent signals of the individual labels have to be singled-out and localized individually. Strategies to isolate single signals include switching the fluorescence of a subset of molecules on and off or from one color to another. This procedure is repeated until the full structure is sampled. The final image is computed from all collected single-molecule positions.
Bioimaging
For super-resolution imaging of living cells a climate chamber is used to simulate physiological conditions with respect to temperature and CO2 levels. The protein of interest may either be expressed as a fusion protein including a fluorescent protein or tag followed by binding of a suitable fluorescently labeled substrate. While it is straightforward to track a single molecule or resolve a static structure in an adherent cell, it is challenging to do the same in an intrinsically highly motile cell, for example in a trypanosome. We employ mechanical immobilization of trypanosomes in a cross-linked hydrogel prior to imaging. The gel has been shown to be cytocompatible on timescales of up to 2 hours and its stiffness may be tuned to achieve immobilization on the nanoscale. We found both the hydrogel and the trypanosomes suitable for single-molecule imaging with respect to autofluorescence levels.
The first structure that we imaged with super-resolution inside a living trypanosome was the axoneme. However, it is our main motivation to gain a better understanding of the structure and dynamics of the trypanosomal plasma membrane including the VSG coat. We used lipid anchored eYFP as a probe to visualize the cytosolic plasma membrane leaflet. Interestingly, this leaflet appeared to be structured. We are currently investigating the potential interaction with microtuble organizing proteins. Next, we will focus on the outer leaflet and the VSG coat itself.
Selected publications
Glogger M., Subota I., Pezzarossa A., Denecke A.-L., Carrington M., Fenz S.F., Engstler M. (2017) Facilitating trypanosome imaging, Experimental Parasitology, doi: 10.1016/ j.exppara.2017.03.010.
Glogger M, Stichler S, Subota I, Bertlein S, Spindler M-C, Tessmar J, Groll J, Engstler M, Fenz S (2017) Live-cell super-resolution imaging of intrinsically fast moving flagellates, Journal of Physics D: Applied Physics, doi: 10.1088/1361-6463/aa54eb
Hartel AJW, Glogger M, Jones NG, Abuillan W, Batram C, Hermann A, Fenz SF, Tanaka M, Engstler M (2016) N-glycosylation enables high lateral mobility of GPI-anchored proteins at a molecular crowding threshold. Nature Communications, doi: 10.1038/ncomms12870.
Beletkaia L, Fenz S, Pomp W, Snaar-Jagalska BE, Hogendoorn PWC, Schmidt T (2016) CXCR4 signaling is controlled by immobilization at the plasma membrane, BBA: Molecular Cell Research, doi: 10.1016/j.bbamcr.2015.12.020
Fenz S, Sachse R, Schmidt T, Kubick S (2014) Cell-Free Synthesis of Membrane Proteins: Tailored Cell Models out of Microsomes, BBA: Biomembranes, doi: 10.1016/j.bbamem.2013.12.009








