Dr. Dominic Helmerich | |
Telefon: | +49 931 31-80449 |
| Since 2023 | Lecturer & Postdoctoral Researcher, Department of of Biophysics & Biotechnology, University of Würzburg, Germany |
| 2022-2023 | Postdoctoral Researcher, Department of of Biophysics & Biotechnology, University of Würzburg, Germany |
| 2017-2022 | Dr. rer. nat., Department of of Biophysics & Biotechnology, University of Würzburg, Germany |
| 2015-2017 | Study of Biology, University of Würzburg, Germany |
| 2012-2015 | Study of Biology; University of Würzburg, Germany |
Single Molecule Localization Microscopy (SMLM) represents a powerful super-resolution microscopy technique that enables the visualization of biological structures at the molecular level.1,2,3,4 This innovative approach transcends the resolution limitations of conventional light microscopy, offering unprecedented insights into cellular processes with remarkable precision.3,4 The fundamental principle of SMLM involves the spatial and temporal separation of emission signals from individual fluorescent molecules. Through sophisticated mathematical approximations of the detected signals, individual emitters can be localized with nanometer-scale precision, facilitating the reconstruction of high-resolution images.1,2
This advanced microscopy method is widely used in many areas of biology, serving as a crucial tool for the detailed analysis of cellular structures and proteins.3 SMLM has established itself as an indispensable instrument in modern biology, and in conjunction with ongoing technological advancements and improved data processing capabilities, it continues to offer significant potential for future discoveries. These developments promise to unveil new insights into the molecular foundations of life, further expanding our understanding of fundamental biological processes.
The remarkable capabilities of SMLM make it a leading microscopic technique, allowing researchers to examine the intricacies of cellular structures and functions with unprecedented clarity. As the field continues to evolve, the integration of SMLM with complementary methodologies and the refinement of associated analytical tools are likely to yield even more significant discoveries about the molecular basis of biological systems.
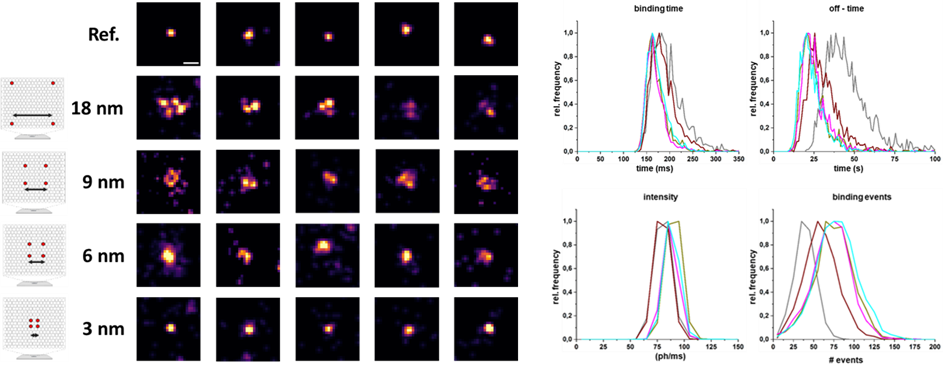
In addition to the single-molecule microscopy method dSTORM, DNA-PAINT represents a powerful super-resolution microscopy technique that enables the visualization of cellular structures with a resolution below 20 nanometers.4 This approach utilizes short DNA strands to label target molecules, along with complementary, dye-labeled DNA strands that transiently bind, resulting in a "blinking" effect.4 The long binding time, in the range of several hundred milliseconds, enables the detection of a multitude of photons, leading to a localization precision in the single-digit nanometer range.
We leverage this high-resolution power to visualize specifically labeled subunits of cellular proteins. Furthermore, we analyze specific oligomerization states, which can be confirmed through specific analyses of binding parameters. While the advantage of this microscopy method lies in its high resolution, it is offset by long acquisition times and high background signal from freely diffusing dye-labeled oligonucleotides, typically restricting its use to Total Internal Reflection Fluorescence Microscopy (TIRF) mode.
Consequently, we are successfully working on reducing the acquisition time of DNA-PAINT measurements and developing techniques to significantly minimize background signal without compromising the high resolution of this method.5,6 These newly developed techniques have also paved the way for volumetric high-resolution imaging of entire cells using DNA-PAINT. This advancement now allows analyses of whole cells with respect to the interaction and aggregation of cell surface proteins
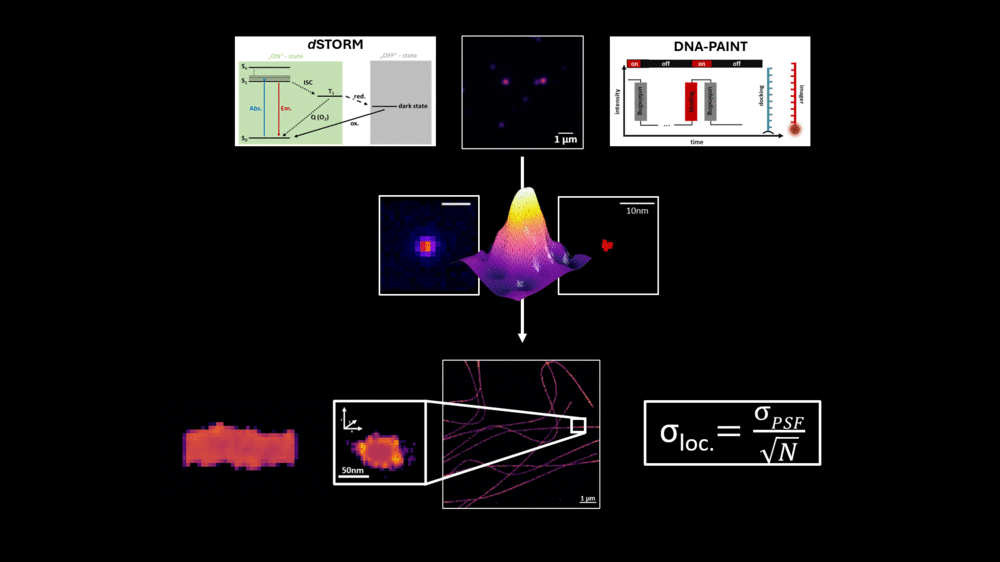
An essential component in evaluating modern fluorescence microscopy methods and approaches is the use of precisely defined reference structures. A widely used tool in this context is DNA origami, a technique that employs DNA as a building block to form arbitrary structures.7 This technique benefits from the ability to accurately calculate and predict specific distances within these DNA-based constructs, making DNA origami a valuable tool in high-resolution fluorescence microscopy.
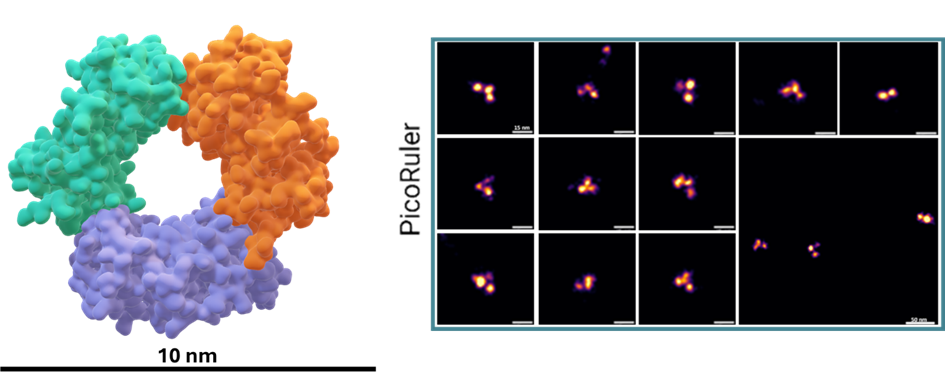
However, in the context of high-resolution biological imaging, DNA origami structures encounter certain limitations. To address this, we have designed a novel protein-based imaging calibration optical ruler (Picoruler) specifically for application in biological systems. These tools are based on the homotrimeric protein PCNA (Proliferating Cell Nuclear Antigen). Through genetic code expansion and click chemistry, these proteins can be precisely modified with fluorescent dyes or other molecules at well-defined positions.8
Our developed Picoruler emphasizes biological compatibility and physiological relevance, as well as stability under cellular conditions. This is combined with high functionality, allowing for modification at precisely defined positions using unnatural amino acids. These properties of the Picoruler were specifically developed for super-resolution biological imaging and calibration at the molecular level.
Furthermore, we are continuously working to expand and improve the portfolio of Picorulers with more complex systems. This ongoing development aims to enhance the capabilities of these molecular rulers, further advancing their utility in super-resolution microscopy and biological imaging.
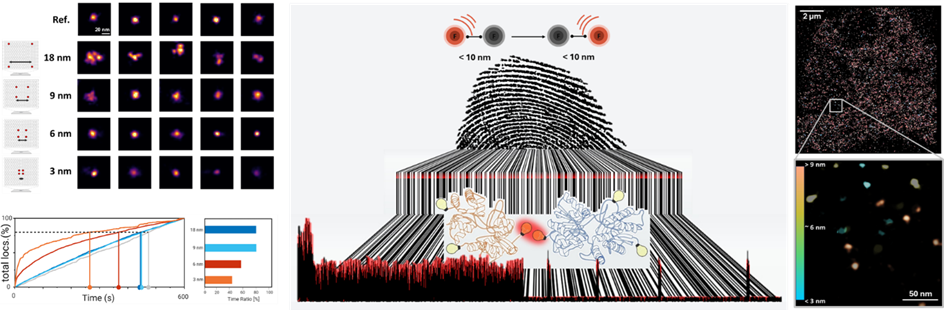
The field of super-resolution fluorescence microscopy is undergoing rapid advancements towards increasing spatial resolution.9 However, this progress is accompanied by escalating demands, complexity, and increased requirements for high technological expertise from users. Consequently, a pertinent question arises: Do established methods and techniques, which are accessible to a broader research community, already include hidden information at the molecular level that can be deciphered through advanced analytical approaches?
To address this question, we employ dSTORM (direct Stochastic Optical Reconstruction Microscopy), a microscopy technique characterized by fluorophore photoswitching, to extract such information. We utilize the characteristic blinking behavior of dyes coupled to target structures to decipher information about distances and numbers of fluorescent molecules in the sub-10 nm range. In combination with genetic code expansion (GCE) and click chemistry, protein structures can be selectively labeled, achieving site-specific marking of the desired structure.8
Our research demonstrates that the blinking behavior of the dyes changes as a function of interfluorophore distance, allowing us to decode a molecular fingerprint of blinking frequency. When used in combination with biocompatible reference structures, this approach enables the elucidation of hidden information at the molecular level. Importantly, this technique is not limited to the class of cyanine dye commonly used under dSTORM conditions and can be broadly applied across various biological research.
This method empowers us to analyze and decipher protein conformational changes or unknown oligomerization states, even when these are not resolvable. The decoded data of characteristic distance-dependent blinking behavior in the sub-10 nm range can also be utilized to unravel different populations of a protein complex on a cell and represent them graphically. This approach thus offers a powerful tool for molecular-scale investigations, enhancing the capabilities of existing super-resolution microscopy techniques.
1) Heilemann, M. et al. Angew. Chem. Int. Ed. 47, 6172-6176 (2008)
2) van de Linde, S. et al. Nat. Protocols (2011)
3) Sauer, M. J. Cell Sci. 126, 3505-3513 (2013)
4) Schnitzbauer, J., Strauss, M., Schlichthaerle, T. et al. Nat Protoc 12, 1198–1228 (2017)
5) Matikonda, S.S., Helmerich, D.A., Meub, M., Beliu, G., Kollmannsberger, P., et al. ACS Central Science 7, 1144-1155 (2021)
6) Ghosh, A., Meub, M., Helmerich, D.A., Weingart, J., Eiring, P., Nerreter, T., et al. Science 387, eadq4510 (2025)
7) Rothemund, P. Nature 440, 297–302 (2006)
8) Kuhlemann A, Beliu G et al. Front Synaptic Neurosci, 13:727406. (2021)
9) Francisco Balzarotti et al., Science 355,606-612(2017)
Helmerich, D.A., Beliu, G., Matikonda, S.S., Schnermann, M.J., Sauer, M. Nature Methods 18, 253-257 (2021)
Matikonda, S.S., Helmerich, D.A., Meub, M., Beliu, G., Kollmannsberger, P., et al. ACS Central Science 7, 1144-1155 (2021)
Helmerich, D.A., Beliu, G., Taban, D., Meub, M., Streit, M., Kuhlemann, A., et al. Nature Methods 19, 986-994 (2022)
Helmerich, D.A., Beliu, G., Sauer, M. ACS nano 14, 12629-12641 (2020)
Kuhlemann, A., Beliu, G., Janzen, D., Petrini, E.M., Taban, D., Helmerich, D.A., et al. Frontiers in Synaptic Neuroscience 13, 727406 (2021)
Helmerich, D.A., Budiarta, M., Taban, D., Doose, S., Beliu, G., Sauer, M. Advanced Materials 36, 2310104 (2024)