Software Toolbox
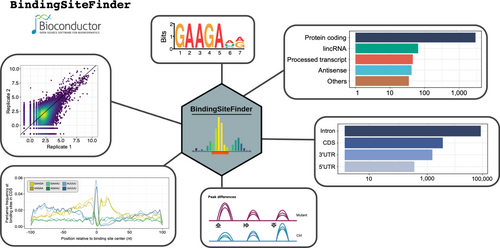
The BindingSiteFinder software package provides a complete workflow for the definition of RBP binding sites. The package provides both functionality and rich visualisations and is well integrated with modern and widely used Bioconductor classes such as GenomicRanges and SummarizedExperiments. The DifferentialBinding module extends the BindingSiteFinder with several new features suitable for identifying RBP-specific binding site changes transcriptome-wide.
Its use is explained in detail in the vignette.
A quick and easy way to visualise and profile high-throughput IP data. This package generates the meta-gene profile and other profiles. These profiles can provide valuable information for understanding IP experiment results.
Their use is explained in detail in the vignette.
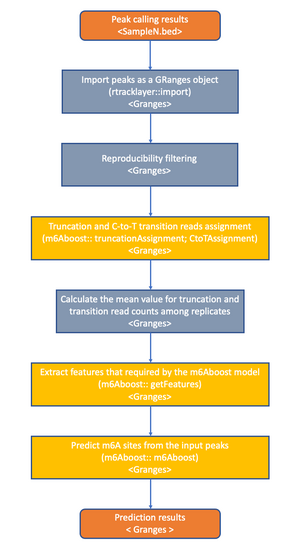
This package can help the user to run the m6Aboost model on his own miCLIP2 data. The package contains functions to assign the siganl and to obtain and use the features of the m6Aboost model.
m6A individual-nucleotide resolution UV crosslinking and immunoprecipitation (miCLIP) and the improved miCLIP2 are m6A antibody-based methods that enable transcriptome-wide mapping of m6A sites at single-nucleotide resolution (Körtel et al. 2021) (Linder et al. 2015). However, due to the limited specificity and high cross-reactivity of m6A antibodies, miCLIP data have a high background signal, which makes it difficult to reliably identify m6A sites from the data.
For accurate detection of m6A sites from miCLIP or miCLIP2 data, we implemented an AdaBoost-based machine learning model (m6Aboost) to classify the miCLIP2 peaks into m6A sites and background signals (Körtel et al. 2021). The model was trained on m6A sites with high confidence obtained by comparing wild-type and Mettl3 knockout mouse embryonic stem cells lacking the major methyltransferase Mettl3. For classification, the m6Aboost model uses a number of features, including the experimental miCLIP2 signal (termination events and C-to-T transitions) as well as the transcript region (5‘UTR, CDS, 3’UTR) and the nucleotide sequence in a 21-NT window around the miCLIP2 peak.
The use is explained in detail in the vignette.
Workflow for analysing CLIP data
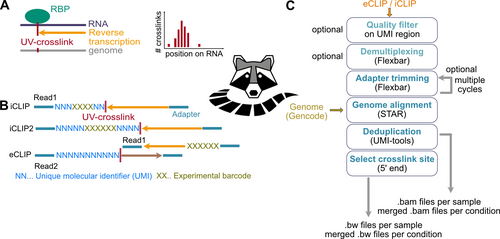
Precise knowledge of the binding sites of an RNA-binding protein (RBP) is key to understanding (post-)transcriptional gene regulation. This information can be obtained from UV cross-linking and immunoprecipitation (iCLIP) experiments with single nucleotide resolution. In Busch et al. 2020, we described the complete bioinformatic data analysis workflow to reliably detect RBP binding sites from iCLIP data. The workflow includes all steps from initial quality control of sequencing data to peak calling and quantification of RBP binding. For each tool, the specific requirements for iCLIP data analysis are explained and optimised parameter settings are suggested. The binding site definition part is implemented in the BindingSiteFinder package.
Simplified use of the workflow with racoon_clip
To simplify the use of the workflow, we have implemented racoon_clip and BindingSiteFinder. racoon_clip is a coomandline tool that automates the processing of CLIP data (Klostermann & Zarnack 2024). It can process both iCLIP and eCLIP data and outputs the crosslink signal with single nucleotide resolution.
The use of racoon_clip is explained in the documentation.
Detecting and analysing circRNAs with Calcifer
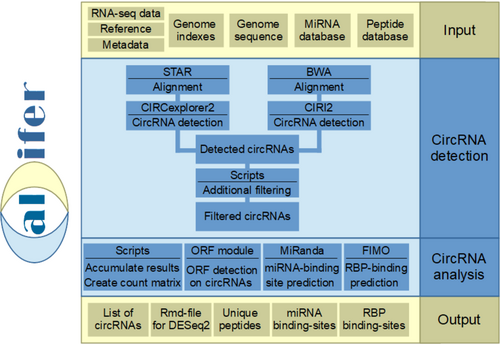
Calcifer is a workflow for the highly automated detection and analysis of circRNAs in RNA-Seq datasets. It enables the analysis of RNA-Seq data for the identification of a list of characterised circRNA isoforms and the prediction of possible functions. Firstly, the circRNAs are detected and filtered from the data. Then the number of reads on the circRNAs and liner RNAs is counted. Finally, miRNA binding sites, RBP binding sites and open reading frames are searched for in the circRNAs.
In Brezski et al 2024, Calcifer was used to compare circRNAs in different subnuclear compartments.
All workflow scripts can be found in the Calcifier GitHub repository.