RNA Splicing in Health and Disease
RNA splicing is one the most important regulatory mechanisms of eukaryotic cells. Deregulated splicing is the cause of various pathological conditions, such as retinitis pigmentosa (RP). Samples of RP patients show a depletion of the ubiquitin-specific protease 39 (USP39).
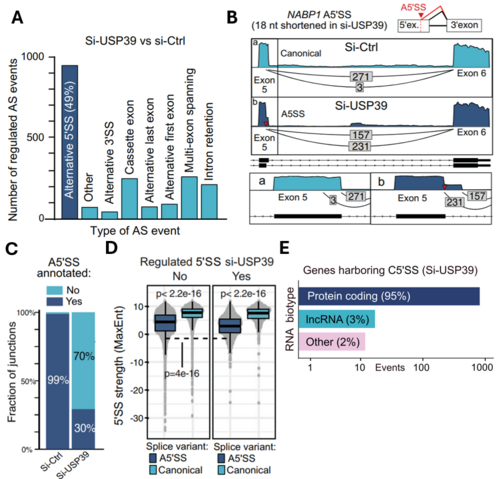
To elucidate the underlying mechanisms of splicing-associated diseases, we analyzed cells with a knockout of USP39. These samples show a strong regulation of alternative 5’ splice sites, which involve an elongation or shortening of the exon up- or downstream of the canonical splice site (A,B). Most of the newly used splice sites are not annotated in the reference genome (C). This so-called cryptic splicing is often caused by an erroneous recognition of suboptimal splice sites. Indeed, the cryptic splice sites used upon USP39 KO are significantly weaker than their canonical counterparts, revealing issues with the recognition of splice sites (D). Additionally, we found that the majority of affected genes are protein-coding (E).
Based on these findings our collaborators were able to show that USP39 increases the aggregation of misfolded proteins by using cryptic splice sites, which eventually leads to cell death.
The full results of this project were published in Pietro-Gracia et al. 2024 .
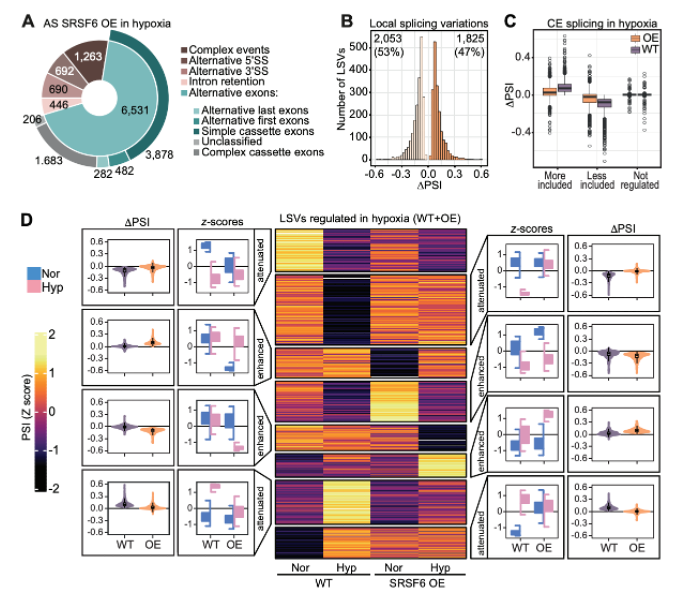
Hypoxia induces massive changes in alternative splicing (AS) to adapt cells to the lack of oxygen. In this project SRSF6 was found to be a key factor in the AS response to hypoxia. A strong reduction in SRSF6 levels in acute hypoxia led to exon skipping and triggered nuclear speckle dispersal as a mechanism for the cell to reprogram the gene expression profile. We found this mechanism to be active in various cancer types and further propose a tumor suppressor function.
In pancreatic β-cells SRSF6 down-regulation promotes β-cell demise through splicing dysregulation of central genes for β-cells function and survival. In this project we characterized the SRSF6 binding landscape in human pancreatic β-cells and identified a purine-rich SRSF6 consensus motif. We also observed, that the positioning of SRSF6 determined the splicing fate by either promoting exon skipping or exon inclusion. Our results also show that such splicing changes can be modulated by the use of antisense oligonucleotides.
The full story was published in de Oliveira Freitas Machado et al., 2023 and Alvelos et al. 2020.