Cytoplasmic RNA Regulation
Spontaneous de novo mutations of the PURA gene lead to a neurodevelopmental disease known as PURA Syndrome. Despite its significance, our understanding of the molecular pathways affected by PURA in healthy cells and its association with these diseases remains limited.
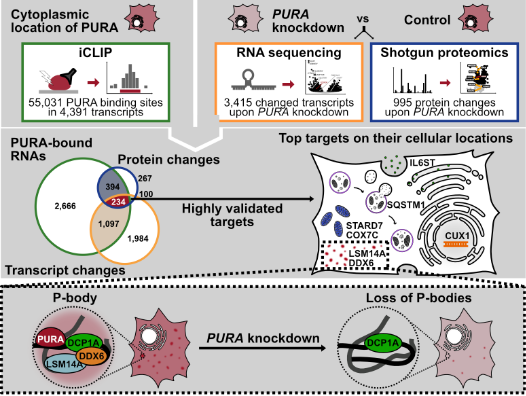
To bridge this knowledge gap, we teamed up with the groups of Professor Dr. Dierk Niessing at Universität Ulm and Helmholtz Zentrum München. Our primary objective is to shed light on the cellular function of PURA in healthy cells, an endeavor we have already begun to explore in our publication titled "Depletion of the RNA-binding protein PURA triggers changes in posttranscriptional gene regulation and loss of P-bodies" (Molitor & Klostermann et al. 2023). Where we unveil the predominant cytoplasmic localization of PURA, where it interacts with thousands of mRNAs. Remarkably, the depletion of PURA leads to changes in the abundance of numerous transcripts. Based on the proteins encoded by these transcripts, we propose that PURA plays a pivotal role in immune responses, mitochondrial function, autophagy, and processing (P)-body activity. Notably, decreased levels of PURA impact the expression of essential P-body components, LSM14A and DDX6, consequently influencing the formation and composition of this phase-separated RNA processing machinery in human cells.
Building upon these findings, our next step involves expanding our understanding of the effects of PURA syndrome through the utilization of physiological cell lines and diverse disease models.